Zheng Lab: Nutrient Sensing and Cell Signaling
Networks
PI: Zhi-Liang Zheng
Professor
Department of Biological Sciences
Lehman College
City University of New York
250 Bedford Park Blvd. West
Bronx, NY 10468
Office: 2404 Science Hall
Lab: 2401 Science Hall
(718) 960-6955 (Office)
(718) 960-5741 (Lab)
(718) 960-8236 (Fax)
E-mail: zhiliang.zheng@lehman.cuny.edu
About the PI
- Asst. Prof. (2003-2008), Assoc. Prof. (2009-2016), Prof. (2016-present), Cellular Signaling, CUNY-Lehman College
- Postdoc., 1999-2003, Rho GTPase Signaling, University of California at Riverside
- Postdoc., 1999, Hormone Physiology, MSU-DOE Plant Research Laboratory
- Ph.D., 1999, Flowering Physiology & Biotechnology, Ohio State University
- M.S., 1994, Horticulture, Chinese Academy of Agricultural Sciences
- B.S., 1991, Horticulture, Fujian Agricultural College
Teaching
- BIO 238 Genetics (undergraduate level, 4 credits: 2-hr lecture, 4-hr lab)
- BIO 400 Genomics and Human Health (senior-level undergraduate course to be developed, 4 credits: 4-hr lecture/discussion)
- BIO 501 Topics in Genetics(graduate level, 4 credits: 4-hr lecture)
Research
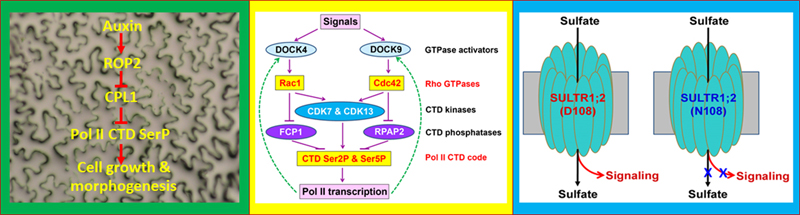
A universal signaling shortcut from Rho GTPases to Pol II “CTD code” modulation in transcriptional control across fungi, plantae, and animalia kingdoms. Rho family small GTPases, including Rho, Rac and Cdc42 subfamilies in yeast and animals and ROP subfamily in plants, are best known as key signaling switches in various cellular activities and developmental processes. Constitutive activation of Arabidopsis ROP2 in transgenic line CA1-1 has been shown to disrupt cell polarity in both leaf pavement cells and root hairs. Our prior forward genetic studies first showed that the CA1-1 enhancer 1 (cae1-1) mutation, which is allelic to MRH2 kinesin gene, converted all root hairs to the bulbous shape in the CA1-1 background (Yang et al., 2007). This finding indicates that MRH2 might be involved in interacting with ROP2 signaling to control the microtubule organization and coordinate with actin filaments.
Genetic and molecular characterization of the second CA1-enhancer (cae2-1) mutant has led us to discover the first transcriptional regulatory mechanism for plant pavement cell shape determination (Zhang et al., 2016). Specifically, we found that cae2-1 is allelic to cpl1 or fry2, indicating that the RNA polymerase II (Pol II) C-terminal domain (CTD) phosphatase CPL1 is a critical transcriptional regulator in ROP2-mediated gene expression. Posttranscriptional modifications in the CTD heptad peptide repeats (Y1S2P3T4S5P6S7), in particular phosphorylation at Ser2 and Ser5, are collectively called the “CTD code” and critical for completing transcriptional cycles for almost all protein-coding genes. Our genetic and biochemical evidence has shown that ROP2 stimulates CTD Ser2 and Ser5 phosphorylation by inhibiting CPL1 phosphatase. Furthermore, we found a similar CTD phosphatase degradation mechanism in yeast Rho GTPase control of cell shape. These results provide convincing evidence that Rho GTPase and Pol II, two key molecules which have been separately studied for more than three decades, are actually linked via the Rho-Pol II CTD code signaling shortcut. This shortcut mode of transcriptional control has an advantage of rapidly bringing about large-scale transcriptional changes (Zheng, 2017). Adopting this transcriptional mode enables eukaryotes to robustly respond to internal cues and external stimuli, and thus increases their environmental fitness.
Recently, we found that the signaling pathway from Rho to Pol II CTD code modulation is also conserved in cultured human cancer cells (Zhang et al., 2020). Using inhibitors and siRNA-based knockdown of Rac1 and Cdc42 GTPases, we have shown that the human CTD Ser2 and Ser5 phosphorylation level is reduced by inactivation of Rac1 and Cdc42 GTPases, concomitant with a reduction of CTD kinases (CDK7 and CDK13) and CTD phosphatases (FCP1 for Rac1, and RPAP2 for Cdc42). Together with our findings in plants and yeast, this suggests a universal Rho-Pol II signaling shortcut evolutionarily conserved across three kingdoms, Fungi, Plantae and Animalia. In addition, we found that the protein degradation inhibitor MG132 reversed the effect of THZ1, a CDK7 inhibitor which could decrease the cell number and the amount of CDK7 and CDK13, accompanied by reduction in the level of CTD Ser2 and Ser5 phosphorylation and DOCK4 and DOCK9 (the activators for Rac1 and Cdc42, respectively). Conversely, treatments of Torin1 or serum deprivation, both of which promote protein degradation, could enhance the effect of THZ1, indicating the involvement of protein degradation in controlling CDK7 and CDK13. Thus, our finding also forms an important basis for developing a potential synthetic-lethal therapeutic strategy that targets the activity or degradation of CDK7 and other protein components involved in the Rho GTPase signaling shortcut model we have proposed.
C, N and S nutrient cross-talk and S sensing in Arabidopsis. Eukaryotes need to respond to dynamic internal cues and external stimuli in order to quickly adapt to their surrounding environments (such as light, temperature, water, CO2 and nutrients). Our research is currently focused on the major nutrients, such as carbon (C), nitrogen (N) and sulfur (S), with a goal of constructing nutrient perception and signal transduction networks. First, through a C, N and S combinatorial design, we revealed that activation of a vacuolar sulfate transporter gene (SULTR4;2) and a putative ?-glucosidase 28 (BGLU28) gene by S deficiency is primarily dependent on the C availability which interacts synergistically with N (Dan et al., 2007). This demonstrates the differential effects of C, N and S nutrients on gene expression. Second, we addressed the C/N nutrient balance signaling by genetic analysis. Cellular C and N metabolism must be tightly coordinated to sustain optimal plant growth and development at the molecular and whole plant systems levels. Furthermore, C/N balance is also critical for the ecosystem response to elevated atmospheric CO2. Despite numerous physiological and molecular studies in C/N balance or ratio response, very few genes have been shown to play important roles in C/N balance signaling (Zheng 2009). Using a genetic approach, we have identified a novel gene (OVERSENSITIVE TO SUGAR1, OSU1) involved in C/N balance response in Arabidopsis thaliana (Gao et al., 2008). Mutations in the OSU1 gene result in the hypersensitivity of the seedlings to the imbalanced C/N (high C/low N, and low C/high N), but the osu1 mutants respond normally as wild-type under the balanced C/N, low C/low N and high C/high N. OSU1 encodes a putative AdoMet-dependent methyltransferase. Interestingly, osu1 mutants are allelic to qua2/tsd2, the cell-adhesion-defective mutants reported by other groups. This indicates that OSU1/QUA2/TSD2 might either have distinct substrates in the control of cell adhesion and C/N balance response or is important in linking cell wall biogenesis and C/N balance response.
In addition, we have taken advantage of the BGLU28:GUS reporter-based nutrient regulatory pattern to isolate S nutrient sensors or signaling proteins. We identified two novel alleles (sel1-15 and sel1-16) of the Arabidopsis high affinity transporter SULTR1;2 in which sulfate uptake was inhibited and gene expression was enhanced (Zhang et al., 2014). Furthermore, we found that the sensitivity in sulfur-induced down-regulation for several genes known to affect S nutrient response was reduced in sel1-15 and sel1-16 alleles even if the internal S status was similar between wild-type and the mutant alleles. This genetic evidence indicates that SULTR1;2 has a dual role in sulfate transport and sensing, which may be classified as a transporting receptor or "transceptor". We are currently working on the mechanism by which SULTR1;2 senses the sulfur nutrient status and how this signal is transmitted to the nucleus. One of the intriguing features of SULTR1;2 is that G208, which is mutated to D208 in sel1-16, is located on transmembrane helix 5 (TM5) and is highly conserved among all transporters related to SULTR1;2 from plants to yeast and animals (Zheng et al., 2014).
Involvement of phytohormone auxin in cell morphogenesis and sulfate nutrient signaling. Auxin is a classical hormone for plant growth, development and response to environmental stimuli. One of the control mechanisms for auxin is to control gene expression. We have found that auxin has a negative regulatory role in Arabidopsis sulfate deficiency response (Dan et al., 2007). Recently, we showed that auxin treatment increased the Ser5 and Ser2 phoshorylation level of the Pol II CTD (Zhang et al., 2016). Interestingly, the stimulation is dependent on ROP2 and ROP4 GTPase activity. This work suggests that the Rho GTPase control of gene expression is important for cell growth and morphogenesis.
Imaging epigenetic studies of drug addiction in humans. Addiction of drugs, including alcohol, cannabis and nicotine, is a heritable brain disease with huge public health, social and economic impacts. Through collaboration, the imaging genetic approach has been used to reveal epigenetic variants involved in alcohol use disorder (AUD). First, it was shown that AUD diagnosis status impacts the correlation between the insular cortical thickness and neuroticism, an important personality trait (Zhao et al., 2017). Subsequently, by using blood DNA methylation profiling, a total of 44 methylation probes, which represent 36 protein-coding genes, are correlated with the insular surface area and significantly associated with the AUD status (Zhao et al., 2019). Among those genes, almost half have been reported to be alcohol-related in humans, including a clock gene PER2 and a taste receptor gene TAS2R16. Interestingly, the other 20 genes might represent novel candidates related to AUD or alcohol response. One of these genes, ARHGAP22, which encodes a Rho GTPase regulator, was hypomethylated in AUD subjects compared to healthy controls, indicating that this Rho GTPase signaling-related gene may be a potential candidate for follow-up studies in the future.
Selected
Publications
- [18] Zhang B, Zhong X, Sauane M, Zhao Y, Zheng Z-L (2020) Modulation of the Pol II CTD phosphorylation code by Rac1 and Cdc42 small GTPases in cultured human cancer cells and its implication for developing a synthetic-lethal cancer therapy. Cells 9: 621
- [17] Zhao Y, Ge Y, Zheng Z-L (2019) Brain imaging-guided analysis reveals DNA methylation profiles correlated with insular surface area and alcohol use disorder. Alcoholism: Clinical & Experimental Research 43: 628-639
- [16] Josh NC, Meyer AJ , Bangash SAK, Zheng Z-L, Leustek T (2019) Arabidopsis γ-glutamylcyclotransferase affects glutathione content and root system architecture during sulfur starvation. New Phytologist 221: 1387-1397
- [15] Zheng Z-L (2017) as and Rho GTPase modulation of Pol II transcription: A shortcut model revisited. Transcription 8: 268-274
- [14] Zhang B, Yang G, Chen Y, Zhao Y, Gao P, Liu B, Wang H and Zheng Z-L (2016) C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast.
Proc. Natl. Acad. Sci. USA 113: E8197–E8206
- [13] Zheng Z-L, Zhang B and Leustek T (2014) Transceptors at the boundary of nutrient transporters and receptors: A new role for Arabidopsis SULTR1;2 in sulfur sensing. Frontiers in Plant Science 5:710
- [12] Zhang B, Pasini R, Dan H, Joshi N, Zhao Y, Leustek T and Zheng Z-L (2014) Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant Journal 77:185-197
- [11] Zheng Z-L (2009) Carbon and nitrogen nutrient balance signaling in plants. Plant Signaling & Behavior 4: 584-591
- [10] Xin Z, Wang A, Yang G, Gao P and Zheng Z-L (2009) The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiology 149: 434-444
- [9] Gao P, Xin Z and Zheng Z-L (2008) The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. Plos One 3: e1387
- [8] Dan H, Yang G and Zheng Z-L (2007) A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Molecular Biology 63: 221-235
- [7] Yang G, Gao P, Zhang H, Huang S and Zheng Z-L (2007) A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 GTPase in Arabidopsis. Plos One 2: e1074
- [6] Zheng Z-L, Yang Z, Jang J-C and Metzger JD (2006) Phytochromes A1 and B1 have distinct functions in the photoperiodic control of flowering in the obligate long-day plant Nicotiana sylvestris. Plant, Cell and Environment 29:1673-1685
- [5] Xin Z, Zhao Y and Zheng Z-L (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiology 139: 1350-1365
- [4] Fu Y, Gu Y, Zheng Z-L, Wasteneys G and Yang Z (2005)
Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis.
Cell 120: 687-700
- [3] Zheng Z-L, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Shen J, Schroeder JI and Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787-2797
- [2] Li H, Shen J, Zheng Z-L, Lin Y and Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiology 126: 670-684
- [1] Zheng Z-L, Yang Z, Jang J-C and Metzger JD (2001) Modification of plant architecture in chrysanthemum by ectopic expression of the tobacco phytochrome B1 gene. J. Amer. Soc. Hort. Sci.126: 19-26 (Won the American Society for Horticultural Sciences Most Outstanding Publication Award of 2001)